DArTreseq
Cost-effective and detailed genome profile analysis technology for any organism.
Detailed genome analysis services
What is DArTreseq?
DArTreseq® (patent pending) represents a novel way of acquiring a very detailed genome profile in a more cost-effective manner than the current “whole genome” sequencing methods.
It extends DArT’s technology portfolio into the realm of enabling identification of the specific sequence features (e.g. Functional SNPs) responsible for an organism’s phenotypes. Our other technologies (DArTseq® and targeted genotyping technologies) work mostly through the detection of marker associations with the phenotype through linkage disequilibrium.
The principle of DArTreseq® is based on effective depletion of repetitive sequences, as well as dramatic enrichment of genic sequences.
Application
While DArTreseq can be applied to any organism, the technology is designed and optimised for plants, as the method takes advantage of the specific methylation pattern of those genomes.
We also recommend the application of DArTreseq to larger genomes, as the cost advantage over the standard resequencing approach is proportional to the size of the genome.
Biochemical removal of repeated sequences and enrichment of genic sequences
The key element of DArTreseq® is biochemical removal of the methylated sequences from the libraries prior to sequencing. This proprietary step has been optimised during our technology development process, in which the DArT team evaluated a number of enzymes which specifically target methylated DNA for degradation.
DArTreseq applies the best enzyme, in optimal conditions, for a specific material presented for analysis.
Enrichment and library construction
Enzymatic treatment is followed by the enrichment step for “low copy” DNA fragments, which are then used in constructing libraries for sequencing.
Based on our experience with different library construction methods in previous projects, DArT will select the optimal method for a specific project or service. All aspects of library construction, from adjusting the DNA concentration to the final step of sequencing DNA fragments, dramatically enriched for low copy sequences, are done in an automated fashion, using modern robotics controlled from start to finish by our Laboratory Information Management System (LIMS) – DArTdb.
Sequencing
Once constructed, the libraries are sequenced using one of the next generation sequencing platforms. We will recommend the optimal platform and sequencing volume based on the intended application of the data and the budget for the project, and after consultation with the potential users of the platform.
Data analytics
DArT has developed a fully integrated data analytics process, connected with our laboratory information management system, so that there is no risk of sample tracking mistakes or data corruption.
This process involves the use of a barcoding system, initially embedded during the library construction step, and carried through to the parsing of individual sequences coming out of the sequencers. This system also uses optimised parameters to align with the sample’s reference genome. It is also possible, at a small additional fee, to align markers discovered with several different reference genome sequences if required.
Data extraction
The final step involves marker data extraction – using either one or a couple of complementary algorithms.
We supply reports in either a standard VCF format, or in formats similar to those we use for other technologies. This allows easier filtering of marker data based on their metadata, and it allows the visualisation of relevant metadata prior to launching of data analysis.
Further information
More detailed information on our specific methodology can be provided as a part of a service report.
DArTreseq precisely targets genes, and avoids sequencing repetitive sequences (“junk DNA”).
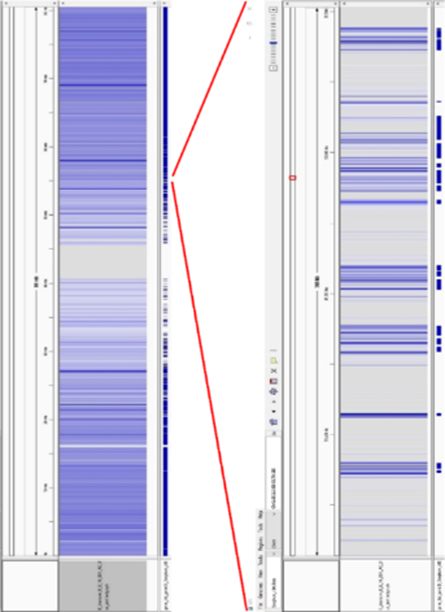
On the left of this image is the overview of chromosome 1 of sorghum.
1 of 3On the right is the zoom into a small segment of this chromosome.
2 of 3Lane 1 in both heat maps shows how many reads aligned to reference – the bluer of the colour the more of the reads are aligned. The light grey colour means no reads. Lane 2 in both heat maps shows the genic regions indicated by blue bars.
3 of 3This picture above demonstrates how efficiently DArTreseq technology enriches for single copy regions, and eliminates intergenic, high copy regions. It is particularly apparent for highly repetitive regions of the centromere.
Related services
Genetic identification
DNA analysis for crops and organisms to identify breeds and varieties.
Quality Assurance Services
Assess your crop, seed, and plant quality for adulteration and purity.