KDDArT platform
Manage phenotypic, genetic and environmental data with our KDDart software platform.
Bioinformatics data integration platform
The KDDart Knowledge Discovery and Delivery System is a data integration platform that provides our clients with the ability to integrate three diverse types of data:
- Phenotypic data
- Genetic data
- Environmental data
KDDart has a flexible design that can be applied to a number of applications – from breeding and pre-breeding activities, to multidisciplinary agro-ecological and environmental research undertakings.
A complete software platform for
geneticists
bioinformaticians
researchers
ecologists
scientists
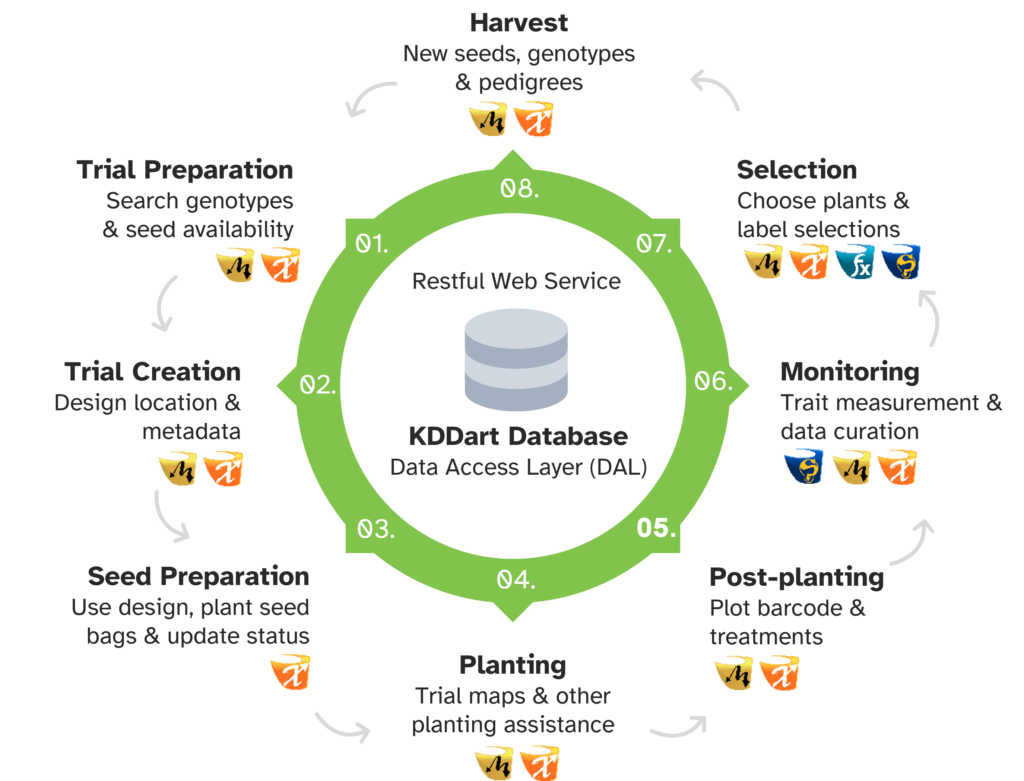
KDDart layered architecture
KDDart was built from the ground up, to open IT industry standards. It consists of a three-layer architecture - an Application Programming Interface between an applications layer and a database layer.
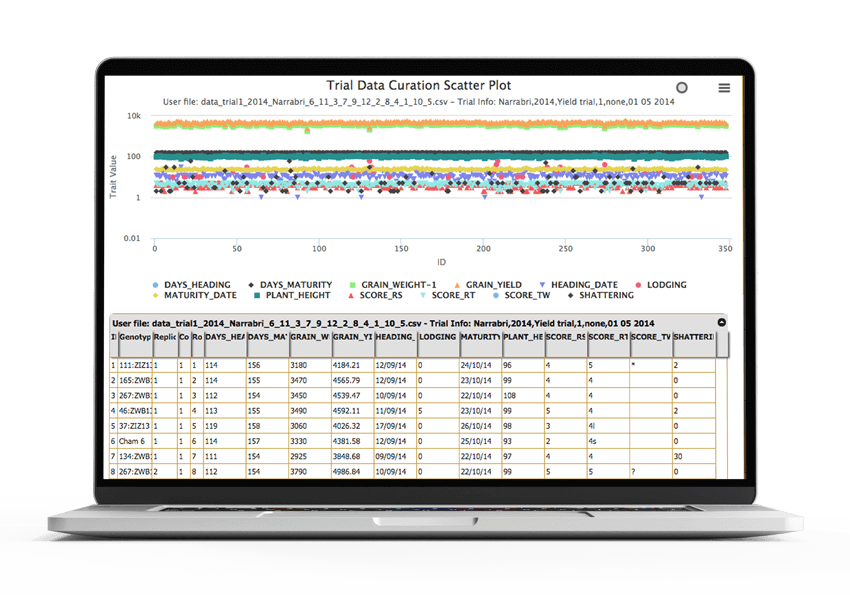
Integrated central storage
The central storage of KDDart is fed from a variety of input media. It, in turn, feeds data out to perform task like analysis, inventory management and preparation for future trials and experiments.
Extensible & flexible
The KDDart infrastructure and applications are specifically designed for extensibility and flexibility to cater for a wide variety of scientific research and experimental activities, with as little restriction as possible.
Applications
KDDart includes five applications that work separately and together to create an integrated solution for your genomic data management needs.
Optimise your research efforts with the power of genetic analysis and big data.
Our full-service bioinformatics and genetic data management platforms provides flexible and extensible IT solutions for a range of genomic, breeding and research applications. Schedule a demonstration today with our team to see how KDDart can support your business.