Targeted genotyping
We offer cost-effective targeted genotyping technology that empowers breeding and research programmes through affordable genetic analysis.
Targeted genotyping services
Overview
Our sequencing technologies DArTseq and DArTseqLD have been used successfully to genetically map hundreds of organisms. These methods are effective in reporting randomly distributed markers.
On many occasions, however, there are applications that require genotyping using only a selected marker set. This is targeted genotyping and we have developed three technologies to achieve these results.
Depending which technology we use there is a variance in their marker density capacity and cost. We will always select the technology for each project based on which is most appropriate and cost effective for each client’s needs. We can offer a cost effective solution to practically any application that needs as few as a dozen and up to around 10,000 markers.
Each Targeted Genotype is developed for the specific material and marker sets required by you. For that reason, this service cannot be ordered through our online ordering system. Please contact us if you want to discuss your needs for specific marker characterisation.
Our Targeted Genotyping uses one of three applications developed by DArT – DArTcap, DArTag and DArTmp.

Our targeted genotyping services help to genetically map a range of organisms.
1 of 1DArTcap
DArTcap applies a selective step after complexity reduction to genotype specific markers from DArTseq representations.
This selection is achieved with the use of the nucleic acid “capture probes” that bind to restriction fragments in the representations carrying the specific DArTseq markers. The restriction fragments are highly enriched in this capture step and most of the sequences obtained from the sequencing of the captured fragments represent the targeted markers.
DArTcap is fairly flexible in terms of the number of markers that can be selected, but in most cases we target from a few hundred to a few thousand markers.
DArTcap is particularly useful in polyploids, as the initial complexity reduction step prior to the capture step would have already selected only one of the homeologous sequences at most loci. This feature enables highly effective sub-genome-specific genotyping as well as obtaining co-dominant SNPs even in highly polyploid species with little or no genome sequence information.
DArTag
Our DArTag method targets a similar number of markers as DArTcap (many hundreds to thousands), but is not restricted to markers identified by the DArTseq platform.
Any SNP (or a small indel) can be targeted using DArTag if there is some genomic sequence available around the variant base/indel.
In the first DArTag step, special molecular probes select the small target regions containing sequence variants. In the second step the targeted regions are amplified and, in parallel, the sample-specific barcode (a short stretch of sequence) is attached.
The libraries generated in that manner are sequenced on the NGS equipment and the resulting sequences processed using DArT’s proprietary pipeline.
DArTmp
DArTmp is used when a relatively small number of markers is required, although it can effectively process a few hundred SNPs in a single assay. We have used DArTmp to deliver as few as 10 markers in one assay.
The advantage of DArTmp is its ability to be increasingly fast as the loci it targets increases. The DArTmp assay is very similar in terms of requirements and the process to the DArTag assay as it also targets a short stretch of sequence around the sequence variant of interest. The main difference is the way the selection of the target sequence happens in the first step of the assay.
In DArTmp a pair of oligonucleotide primers is used instead of a single probe applied in DArTag. All the consecutive steps in the lab and the whole analytical process are the same for DArTmp and DArTag. As DArTmp is targeting the smallest number of markers, the initial investment in assay development and oligo synthesis is small, enabling rapid adoption of this method to a number of applications requiring only a modest number of markers.
Targeted Genotyping Marker Design
DArTag and DArTmp technologies allow for the genotyping of known SNP and small indel markers, with panel sizes ranging from tens to thousands of markers. During the marker design phase, the markers which are to be converted for the targeted genotyping panel will be processed through our proprietary marker design and QC pipeline.
The only essential information required for this is the marker variant information, plus some flanking sequence. The availability of a reference genome assembly is not essential, although the use of a reference assembly can assist in the marker QC and selection process.
A targeted genotyping panel can be designed from your choice of markers.
Additionally, we can convert markers derived from DArTseq and DArTreseq data, providing a very cost effective method for marker discovery in your samples.
A successful targeted genotyping marker panel will match the markers to the material to be genotyped. The DArTag and DArTmp design process gives the flexibility needed to ensure the right combination of markers to avoid ascertainment bias.
The composition of DArTag and DArTmp panels can be updated over time to suit the changing requirements of a genotyping program.
The marker design submission format is described in the document titled Guide For Marker Submission and Specification Form. This document contains the formatting requirements and an explanation of the essential and optional fields in the submission table. This document also provides a form in which to provide instructions on how you would like the marker design and QC to be performed.
An example submission table with descriptive information is available in the document titled Format Instructions For Marker Submission With Examples.
A blank template submission table is also available in the document Template File For Marker Submission.
Targeted Genotyping Workflow
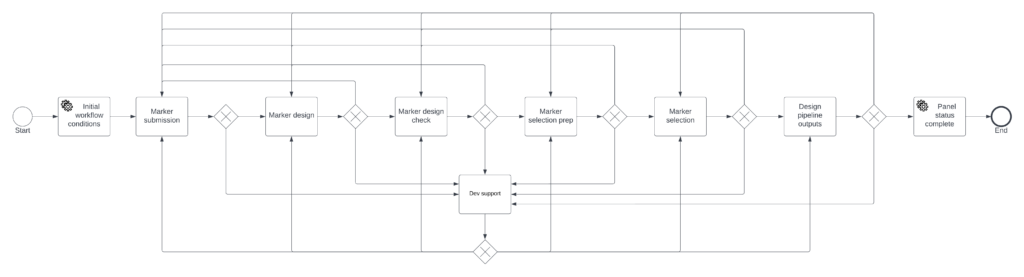
Targeted Genotyping Pipeline
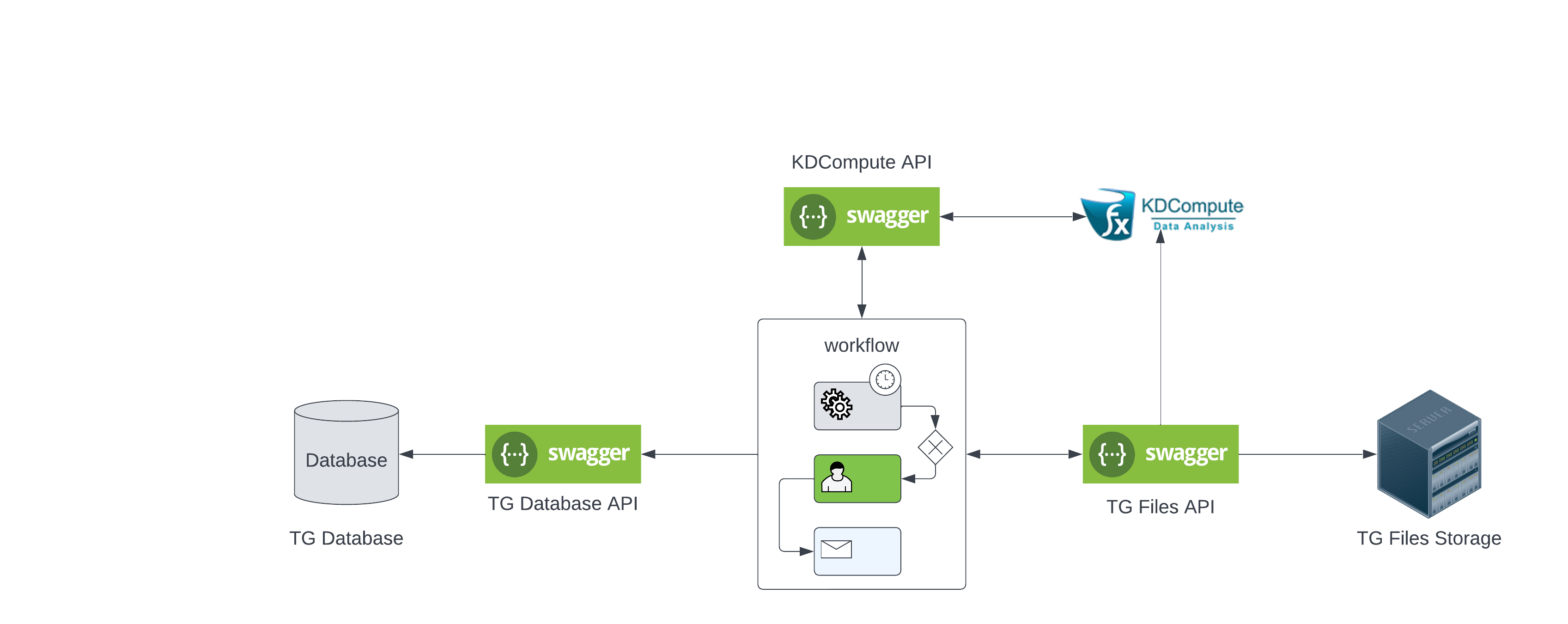
Important information
When ordering Targeted Genotyping (using DArTag or DArTmp) please note:
- DArTmp and DArTag services are processed in 384-well PCR plates, thus we require the services to be ordered in multiples of four 94-well PCR plates. Wells G12 and H12 must be empty for our internal controls.
- We can process partial 384 well plates, however the cost per sample will be higher.
- Both services require the submission of a positive control (PC)
- Minimum 500µl of ~25ng/µl of good quality gDNA in EB buffer (10mM Tris-Cl, pH8.5) shipped in a microtube.
- We recommend checking for presence of residual nucleases in your DNA prep as it may degrade the gDNA during transportation. Follow instructions in the FAQ “How can I check DNA quality for DArT assays?”
- PC can be shipped as precipitated in Ethanol or as dry pellet. Please indicate DNA storage method and gDNA quantity.
- Alternatively, we can extract gDNA for PC in our facilities. Submit enough tissue to obtain 10-12µg of gDNA
- Please include a printed copy of the Positive Control Specification document (download below) and all other required documentation. See our ordering pages for the documentation required for sending tissue or DNA, and from inside or outside Australia.
Downloads
To download the following files, visit our Help Centre.
- Guide For Marker Submission and Specification Form
- Template File For Marker Submission
- Format Instructions For Marker Submission With Examples
- Positive Control Specifications
If you require any additional information about the submission of a Positive Control, please contact us.
Related services
Quality Assurance Services
Assess your crop, seed, and plant quality for adulteration and purity.
Genetic identification
DNA analysis for crops and organisms to identify breeds and varieties.